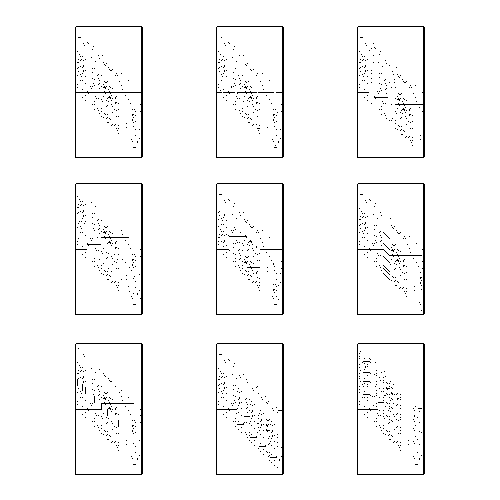 .
.
A sequence is a string of entities from a finite alphabet with some ordering relationship, e.g. text, shapes, music, speech, pictures. Relationships between sequences include equality, containment, cyclicly equal, or partial overlap, along with differences caused by substitutions, insertions, deletions, swaps, compressions, and expansions. Relationships between entities in a sequence can be binary (equal/not equal) or multilevel.
A Correlation Image (CI) is a visual representation of all possible alignments between 2 sequences. For 1-D sequences, each row corresponds to a different alignment of the sequences, and the intensity of each pixel is proportional to the goodness of match of the aligned entities.
The following is a list of perceptible structures within the CI and the generalized sequence relationship which created them.
This image shows examples of each of these phenomena.
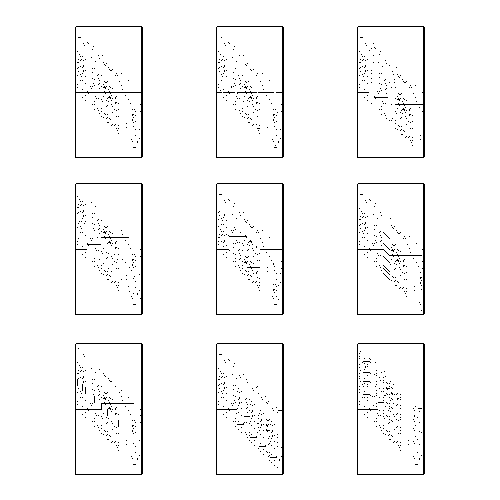 .
.
XSauci (X-window Sequence Analysis using Correlation Images) is a system developed at WPI by David Nedde and Matt Ward (with later enhancements by Maureen Higgins) for exploring the application of CIs to genetic sequence analysis. The following is a partial list of the features of XSauci:
Biologically significant relationships can be localized by searching for the features listed below.
A recent thesis by John Rasku explored the use of CIs in comparing shapes. By representing a shape as a sequence of elements (we tried the coordinates themselves, first and second derivatives, and distance to centroid) with a distance metric, we could classify the way different shape changes manifested themselves within the CI and which representation(s) were best for various shape differences. We did a simple experiment with 3-D shapes as well (represented as a series of 2-D slices), though it was clear that better 3-D representations were needed.
matt@owl.WPI.EDU